abYsis is the world's most widely used antibody workbench, enabling teams world wide to make discoveries faster.
Accelerate your research and discovery with the leading antibody analysis platform. Search the existing database of published sequences, or use flexible input tools to annotate DNA or protein sequences from your own research.
Antibody Analysis
Let abYsis rapidly generate 'antibody aware' multiple sequence alignments and then use annotation to further prioritise.
Explore sequences in depth with Key Annotation. Interrogate CDRs, PTMs, unusual residues, germline hits and more. Assess unusual residues and edit with the Humanize platform.
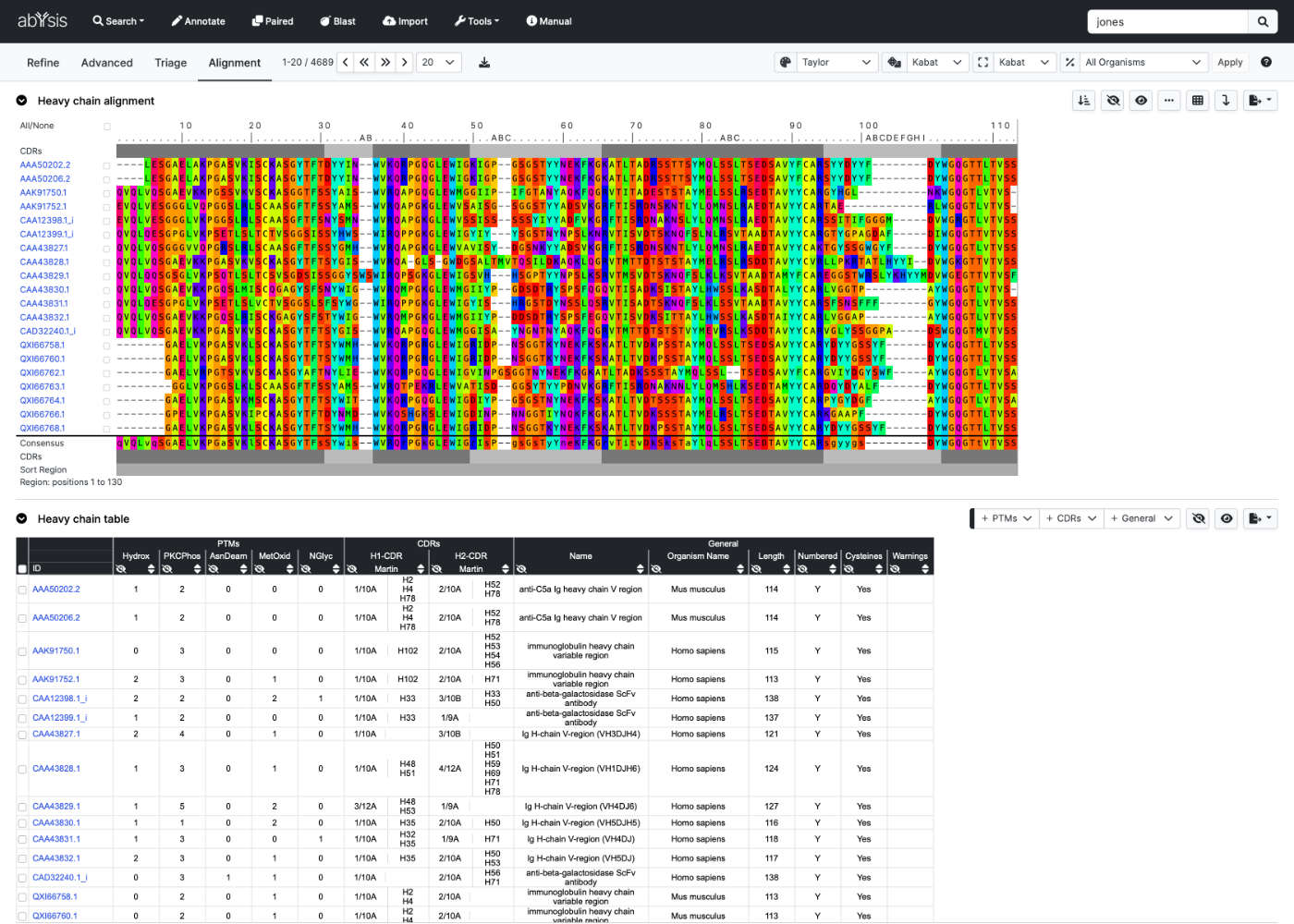
abYsis lets you focus on research
rather than spending time learning how bioinformatics algorithms to work.
Paired analysis
Paired Analysis allows you to analyse heavy and light chains using a tailored version of the Humanize facility.
Either select data from the database or load 2 sequences to Annotate together.
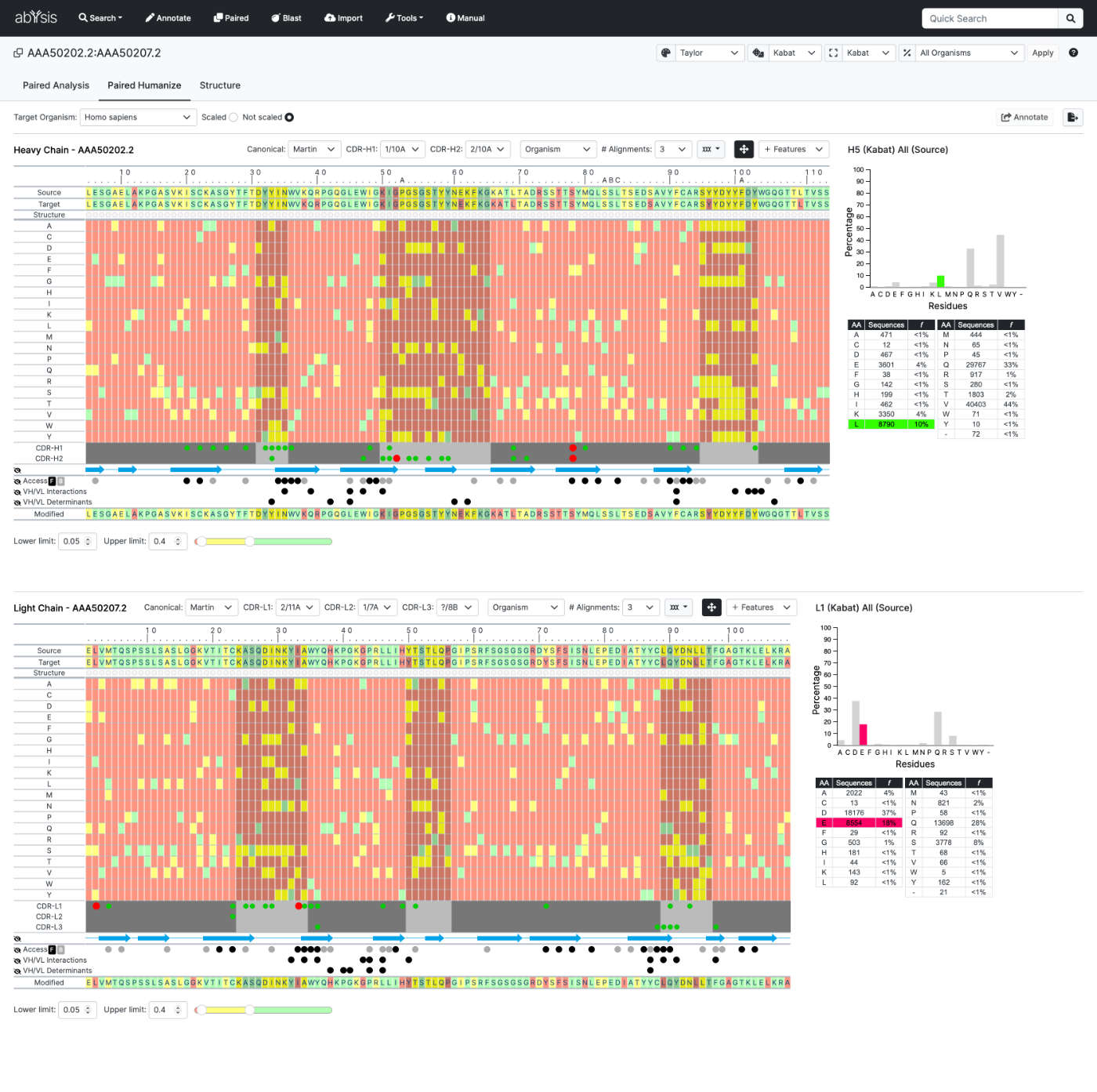
Homology Search
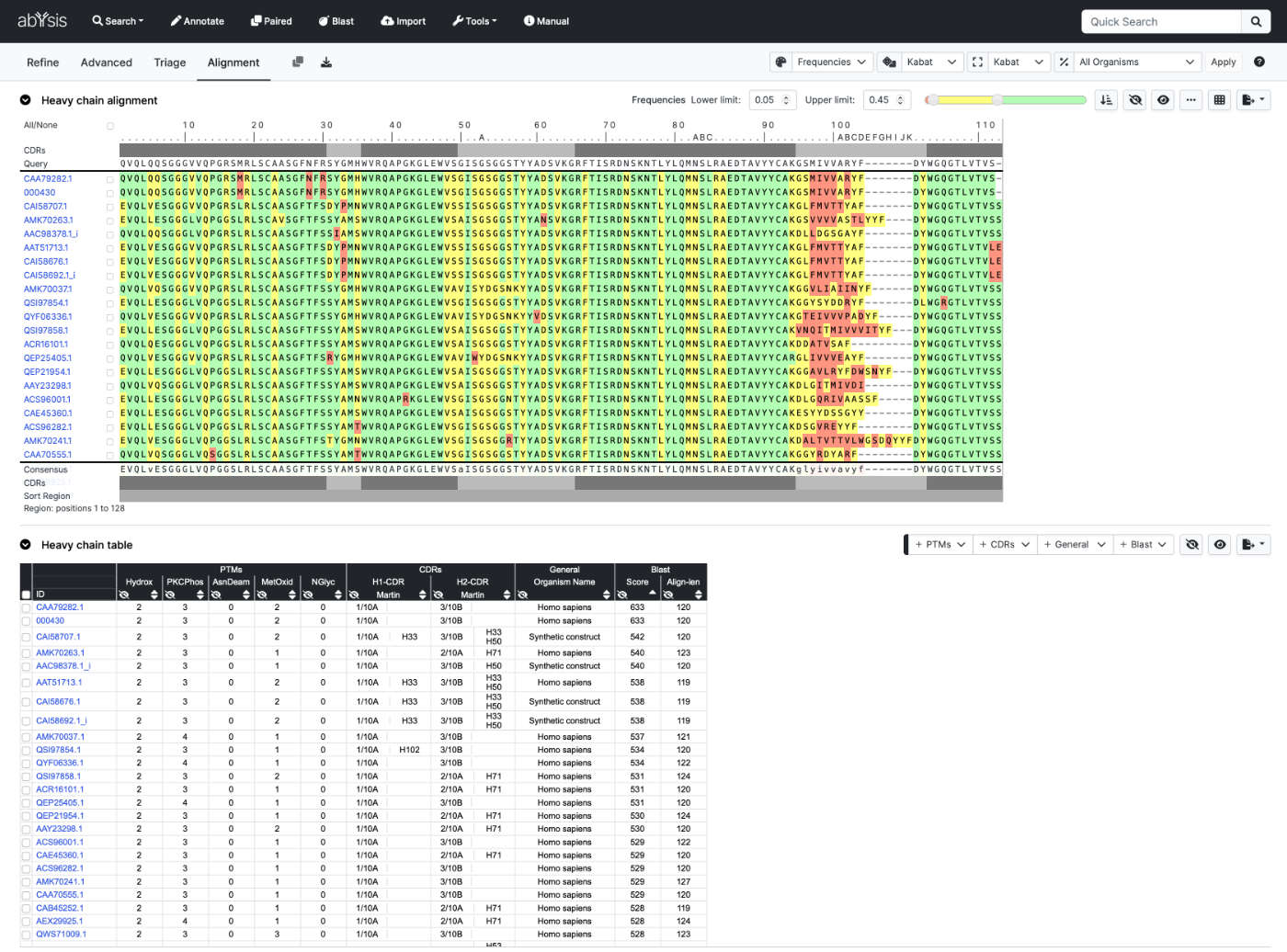
Perform a quick homology search of antibody sequences in abYsis using either DNA or Protein input.
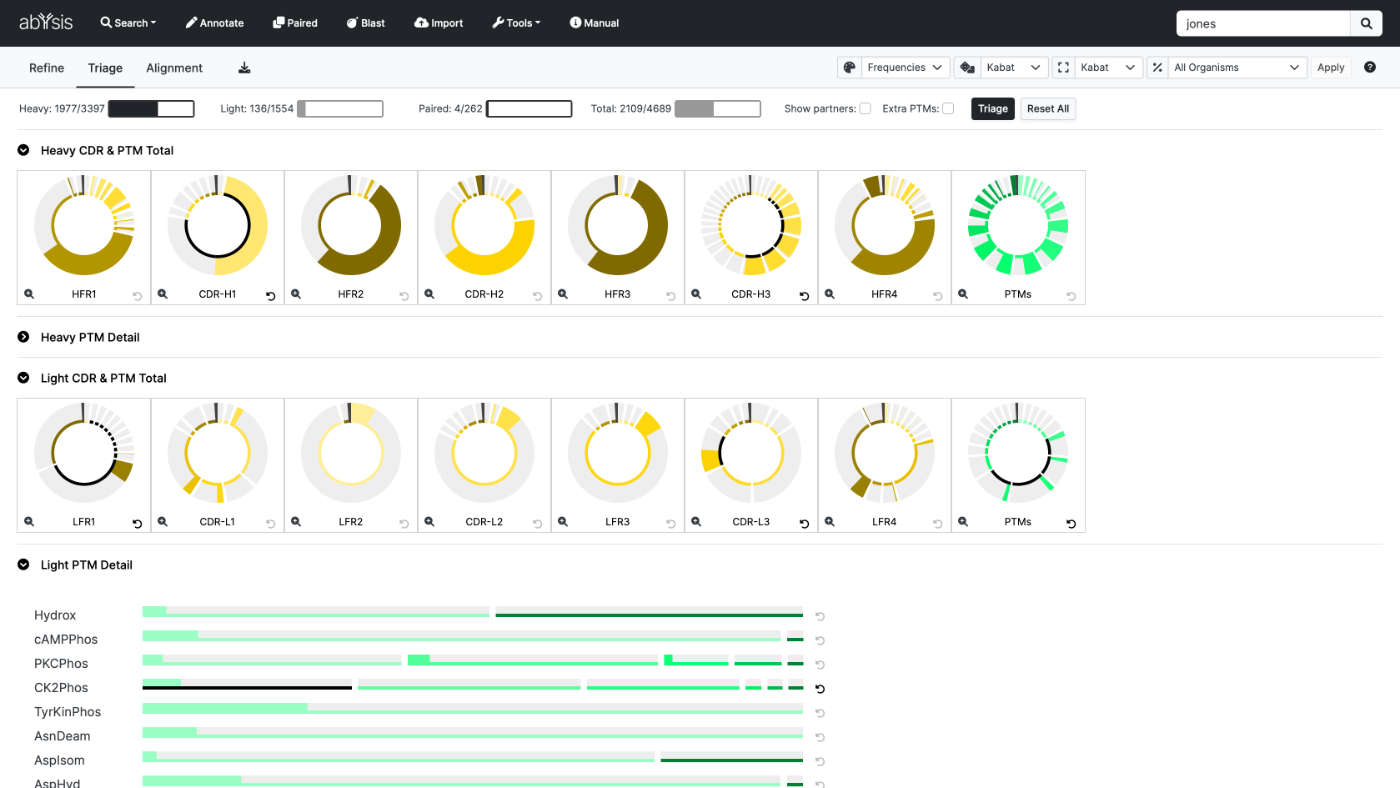
Triage
Benefit from the Triage facility and filter large datasets based on user preference.
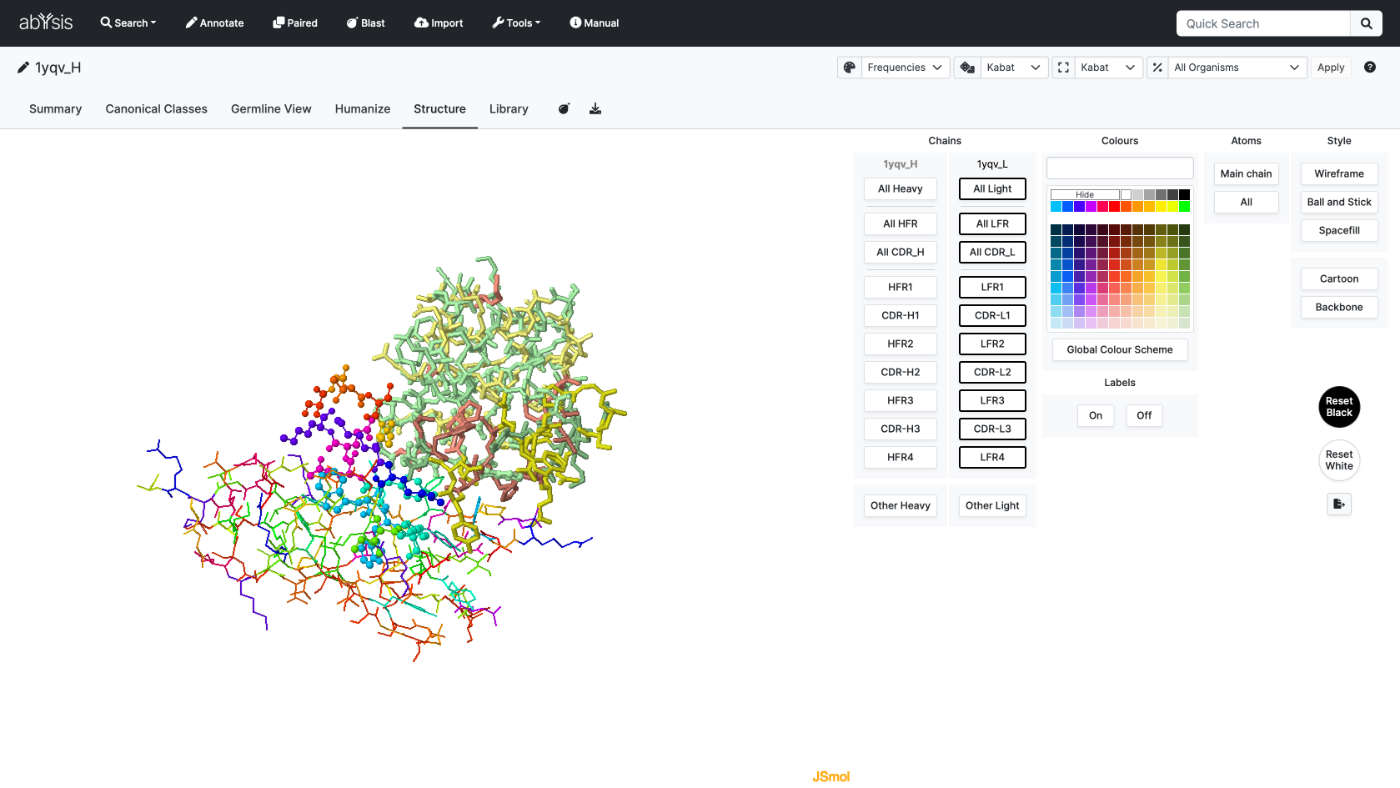
3D Structure
Analyse sequences with 3D structure coordinates. Structures can be either experimental or homology models, from either internal research or a public resource.
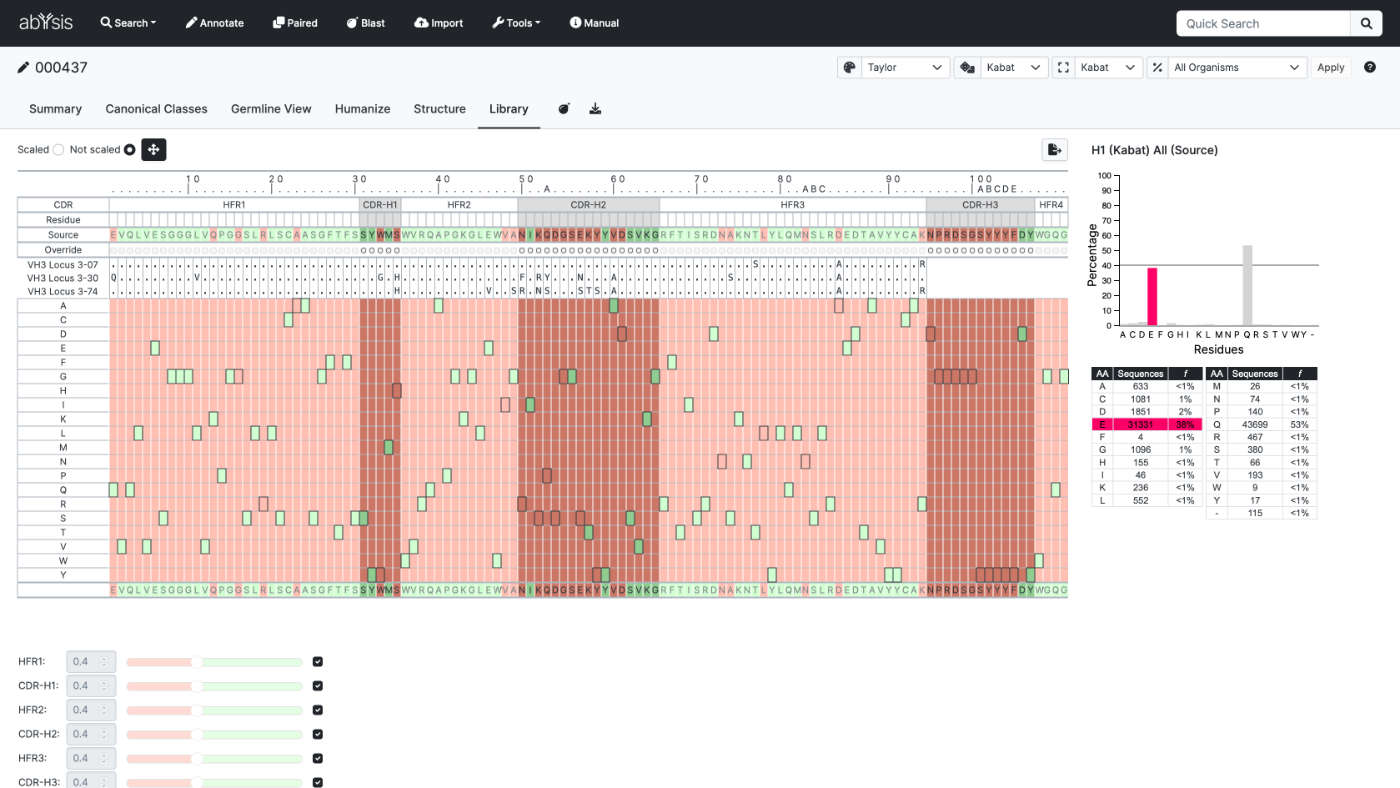
Library
Design your own libraries based on frequency data in abYsis.
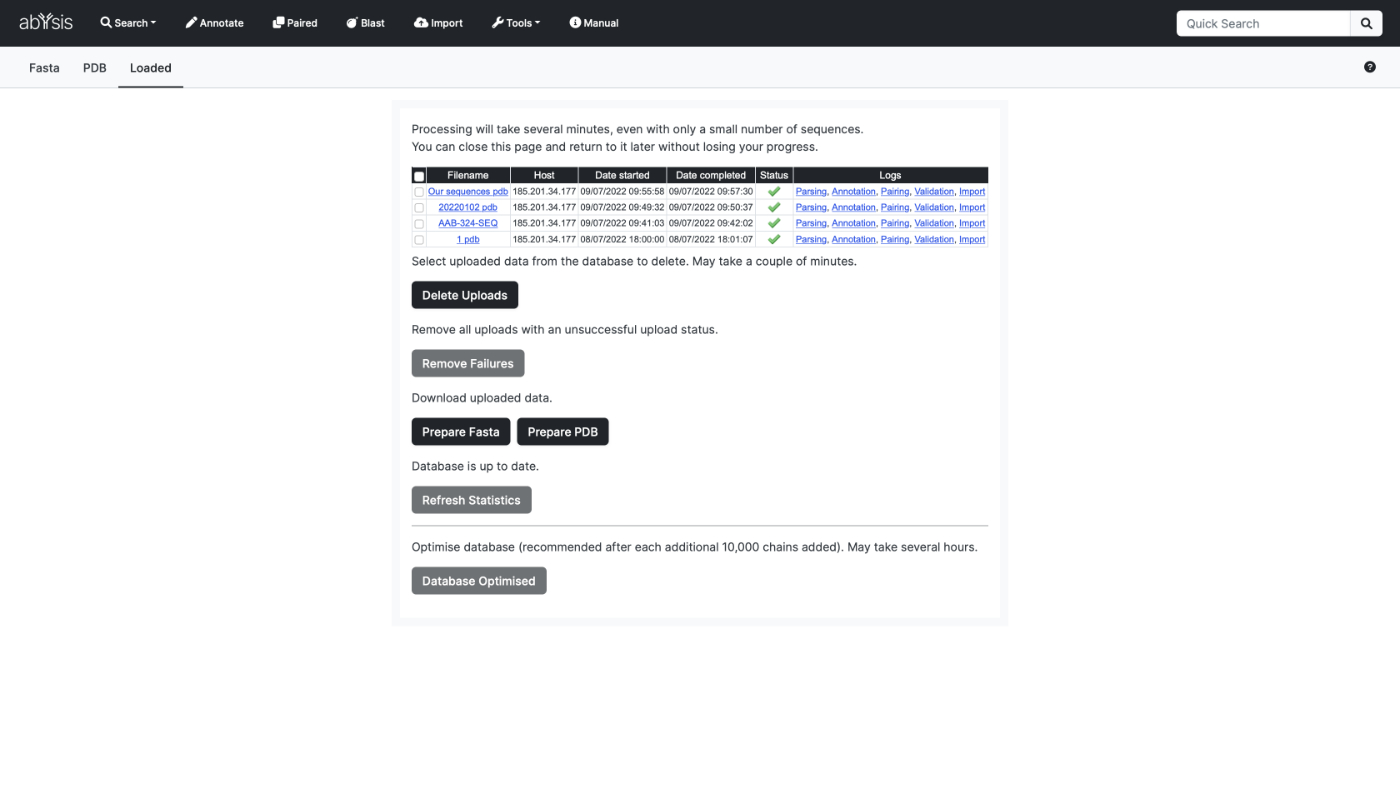
Import
Import your own sequences & structures and store for later use. Use 3D coordinate data from internal projects or updates from the Protein Data Bank.
Key Features
Protein and DNA Input
Use your own protein sequence translations or input DNA and let abYsis search for the appropriate reading frame for translation.
Automated Antibody Numbering
Apply standardised numbering and select from a choice of Kabat, Chothia, Martin, IMGT and Aho methods.
Humanness Assessment
Estimate humanness of sequences under investigation
Framework and CDR Assignments
Select from a choice of Kabat, Chothia, IGMT and other assignment methods best suited to your research.
3D Structure Integration
Combine sequence with 3D structure data
Canonical Class Annotation
Predict canonical class based on sequence using rules derived from well known published methods.
Colour Selection
Colour sequences using either abYsis frequency distributions or converntional property characteristics.
Unusual Residue Identification
Quickly identify statistically unusual residues at key positions of your sequence and decide whether there is benefit to adjusting.
Humanization
Use the heatmap view of unusual residues and compare species differences to help humanize your sequence.
Relational Database Storage
Benefit from storage in a PostgreSQL database. Add further sequence and structure data for flexible analysis
abYsis was developed by Andrew Martin's Bioinformatics and Computational Biology Group at University College London.
