-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
Alignment and Table
Note!
Depending on the query, there can be a difference between Triage and Alignment tabs. Specifically
- Triage tab; Only displays sequences that match the query.
Within those hits it also gives the opportunity to Triage Heavy/Light pairings - Alignment: Displays sequences that match the query
AND paired chains that did not directly hit the query.
Global Settings 
Pagination: For database searches the default is 20 results per page but this can be increased. Results are split into pages and navigation buttons allow the user to progress forwards/backwards by page as well as to go to the first/last page.
- Colours: Select amino acid colour scheme. abYsis frequencies are now available
- Numbering: Select preferred numbering scheme
- CDRs: Select preferred Framework/CDR definition scheme
- % (Amino acid frequency): Select organism on which to calculate abYsis frequencies
Alignment
Alignments are shown in three sections, Heavy and Light as well as Combined Heavy with Light for paired antibodies

Alignment view only shows numbered antibody sequences. Sequences in abYsis that have not been numbered are not displayed.
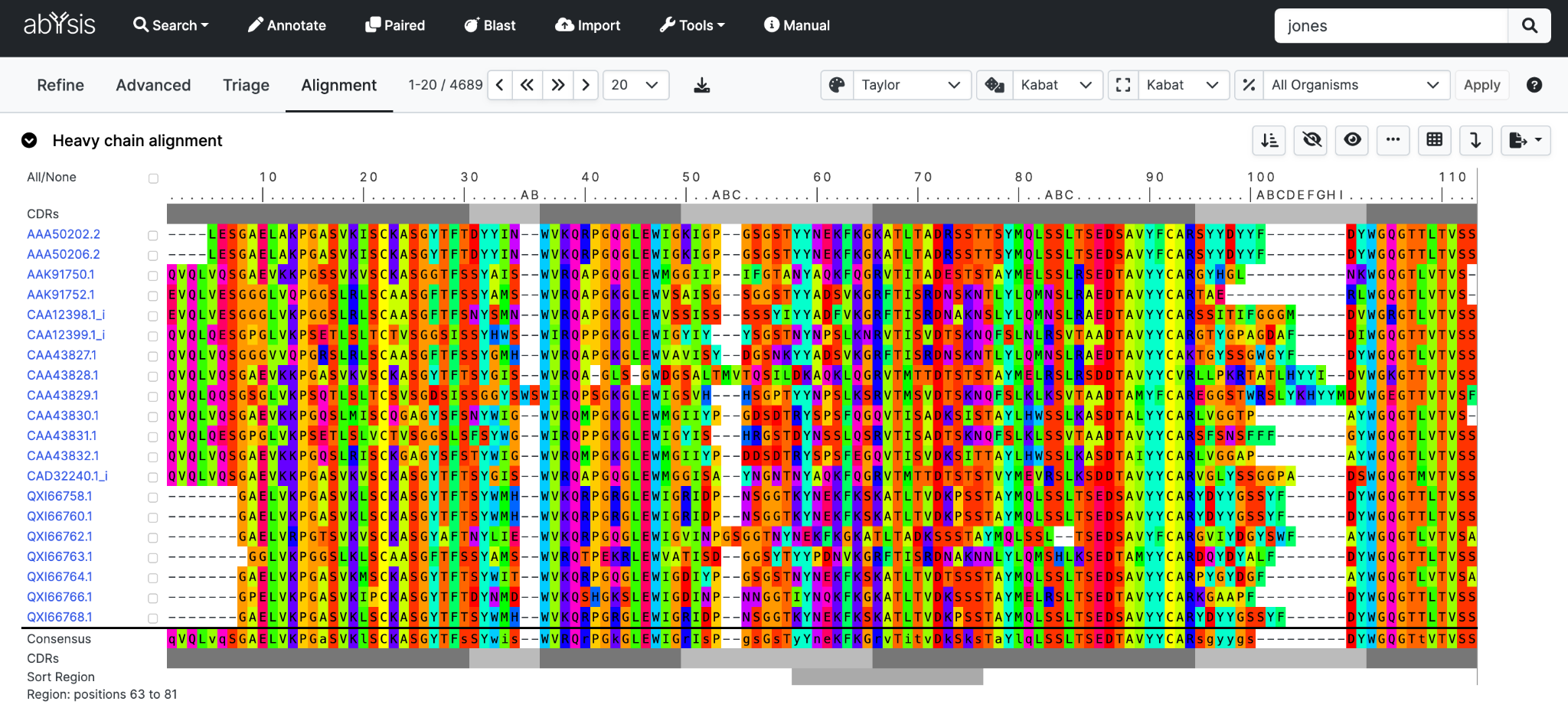
Hovering over the [...] key will show residue numbers.

Below the key and at the bottom of the alignment is a bar indicating the frameworks (dark grey) and CDRs (light grey) using the Global definitions.

A simple Consensus sequence is shown at the bottom of the alignment.
Upper case indicates a residue type found in >50% of displayed sequences where lower case is the most common but does not cover more than half of the sequences.
Sort
Sort Sequences can be quickly sorted based on sequence similarity.
A representative sequence (most similar on average to the other sequences) is placed at the top of the alignment, with the other sequences shown in groups gradually changing from the representative.
By default, sorting is performed over the whole sequence, but the region over which the sort is displayed can be changed using the Sort Region.

Here we show a short sort region covering part of CDR-H2 and FR-H3

Clicking Sort will re-order the alignment and associated data table.
Other Controls

The Dotify checkbox causes residues the same as the residue above in the alignment to be represented by a dot.
The Nocolour checkboxes removes the colouring of those residues. This enables one to focus on differences in the sequences.
Hide Selected sequences By first checking the box next to a sequence identifier clicking the Hide button will remove sequences from view. Sequences will also be hidden in the associated data table.
Show Hidden sequences All hidden sequences will be re-displayed.
Transpose view Re-orient to allow a vertical view. Note that sorting by sort region is not included in this view.
Exporting sequences
Excel format is the most comprehensive and will display both alignment and table together.
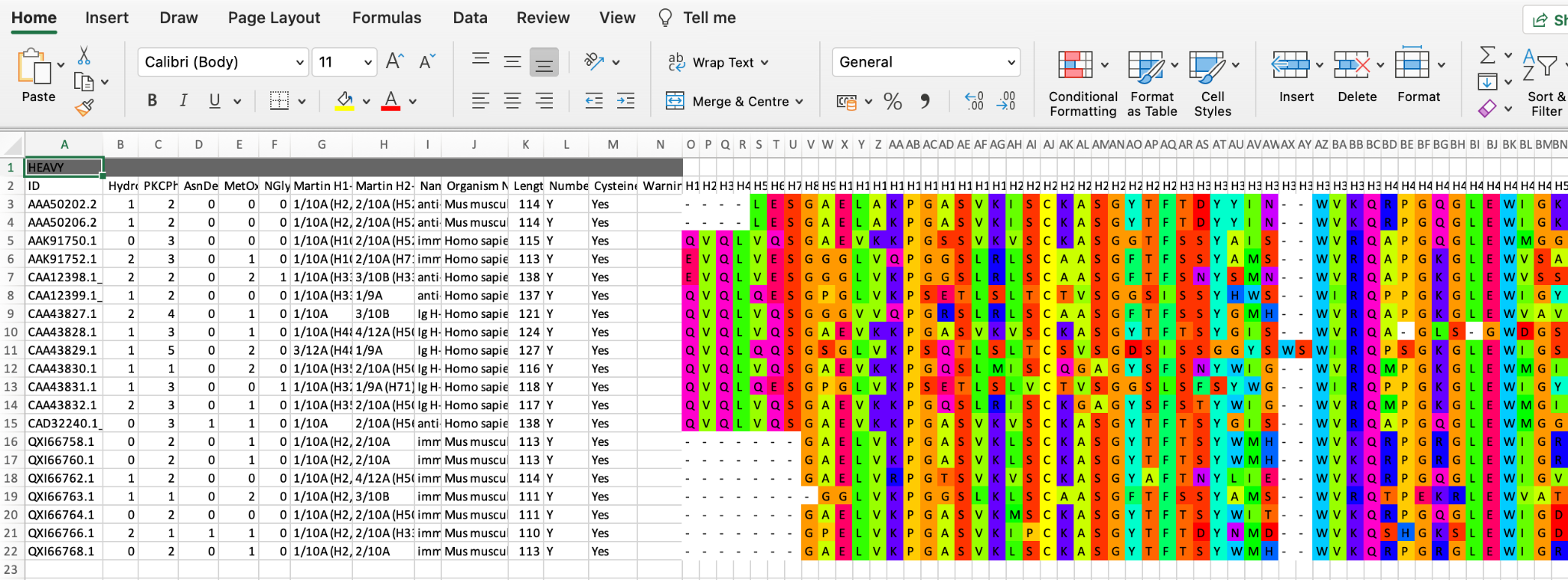
FASTA format. Select the sequences to be exported (selecting no sequences is the same as selecting them all) and click the Export button. These will be shown as an alignment with dashes for gaps

CSV (comma-separated values) format is the same as Excel but without the colouring and formatting.
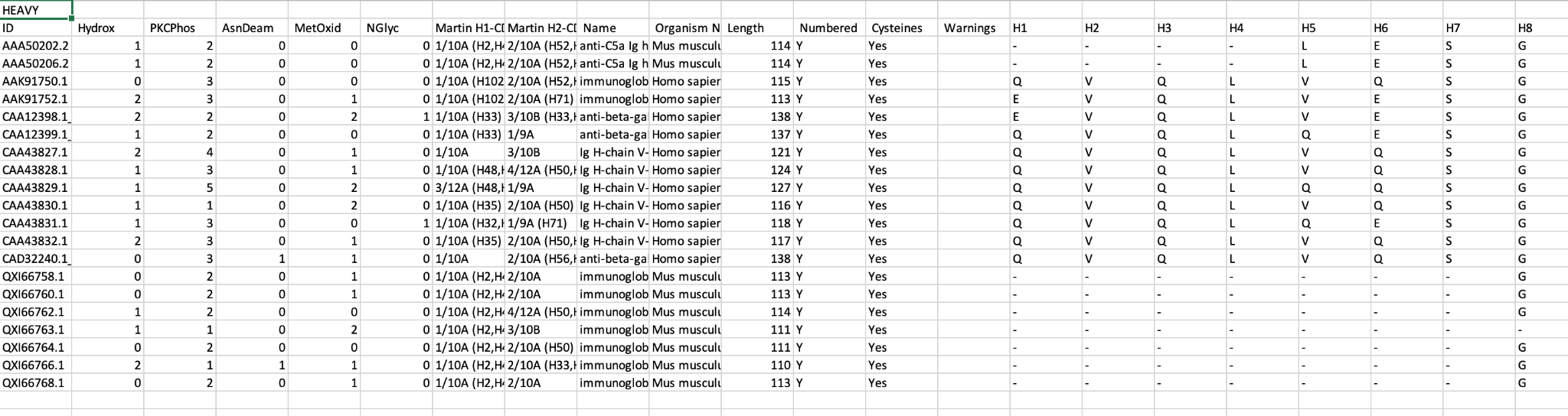
Table
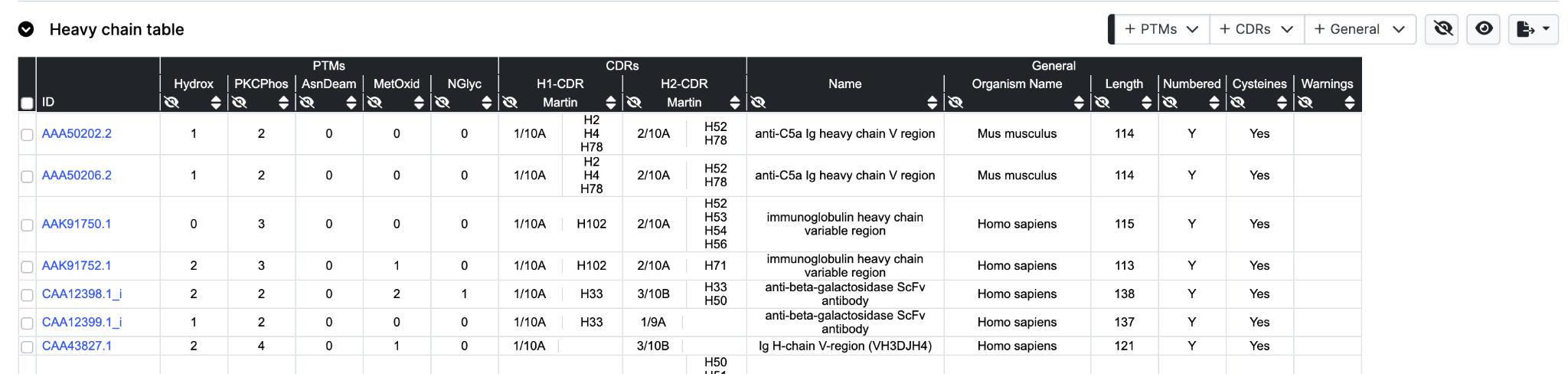
The data table view displays annotation from three dropdown menus, with a selection turned on by default.
Hide works in a different manner to hiding rows. Columns are removed by clicking the respective hide icon to the bottom left of each header row cell.
Sorting and reverse sorting is achieved with the respective sort button at bottom right of each header row cell. Note that the sequence alignment view will also be re-ordered based on the sort.
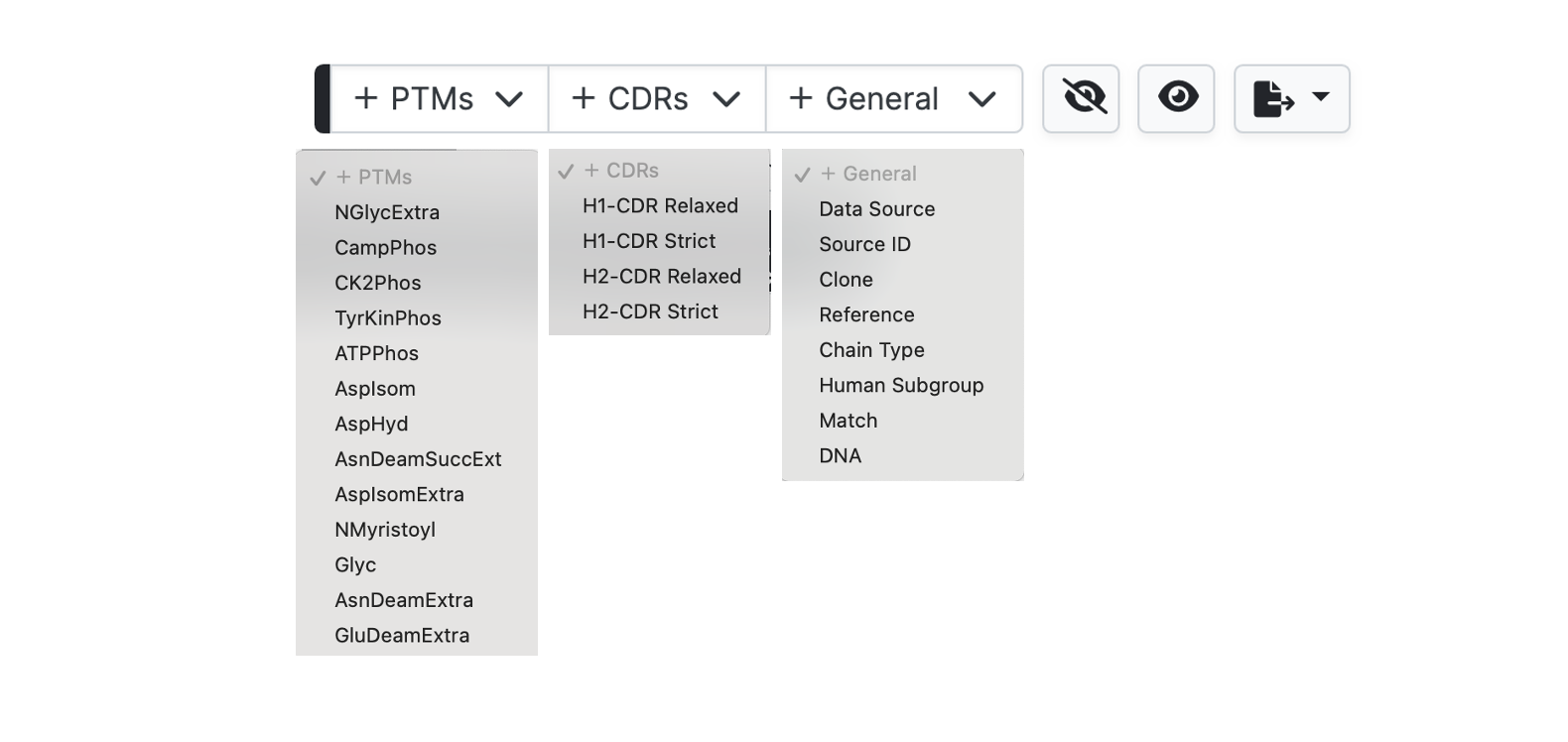
To add further columns to the output simply select from the pulldown menus. Note that for the General tab only Human Subgroup is calculated (where appropriate) by abYsis. Other information is take from the source files.
