-
Overview
-
Sequence Input
-
Database Search
-
Multiple Alignment
-
Key Annotation
-
Structure Input
-
Paired
-
Tools
-
Miscellaneous
-
Statistics
-
Licence File
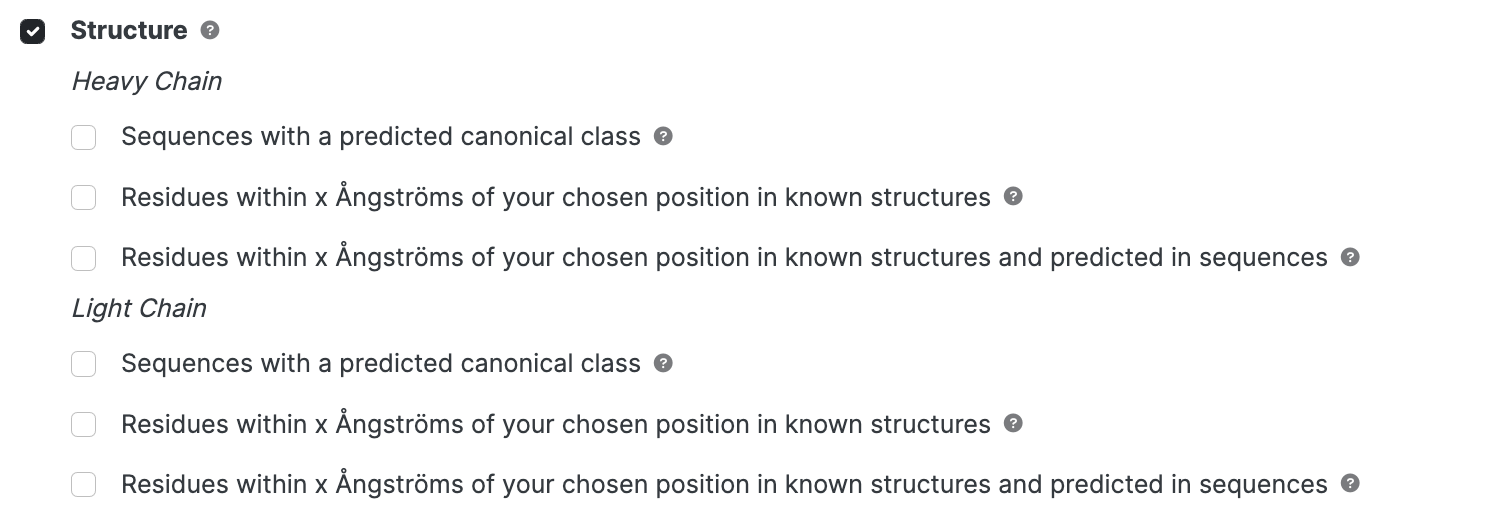
Structure
Structural searches are for advanced users. Some options require structural information and for those, the effect is to limit the search to PDB entries loaded in to abYsis.
Note:
- Search terms may be entered separately for heavy and light chains.
- Specifying both chains in a single search will be restricted to paired chains.
Sequences with a predicted canonical class
Restrict output to CDRs that predicted to belong to a specified canonical class based on the presence of key residues.
Each canonical class (a structural concept first proposed by Chothia) is encoded by rules that require particular residues at specified positions for a match. Using these rules, canonical classes can be predicted for all numbered chains, irrespective of whether structural information is available.
- Choose whether you require an Exact only (meaning exact match to the published residues) or wish also to Allow similar matches.
- Then for each CDR of interest:
- Select the classification method using the Method (Martin, Relaxed or Strict)
- Select the desired canonical class from the Canonical Class dropdown
See the Definitions pages for a description of CDRs and the residues required at each position.
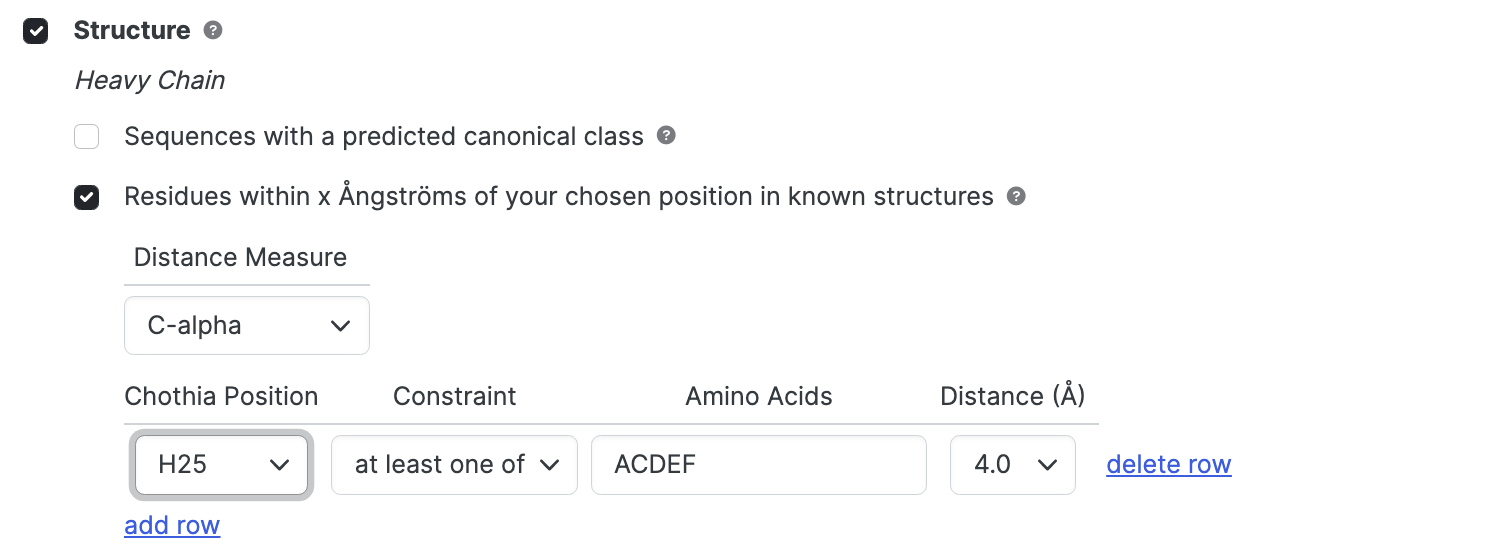
Residues within X Ångströms ; 3D structures only
Restrict the search to structures that have (or do not have) particular amino acid types within a particular distance of a specified residue position.
Note: This will restrict the search to entries with 3D coordinates
- Use the Distance Measure dropdown to specify how the distance between two residues is measured - the distance between C-alphas, or the distance between closest atoms.
- Use the Chothia Position dropdown to select a key position. Note that only chains that have the specified position (and have atom coordinates for that position) will be returned.
- Use the Constraint dropdown to select the type of constraint you wish to apply - either at least one of or none of. This controls whether the specified amino acids should be present or absent.
- Use the Amino Acids box to specify the amino acid types of interest using 1-letter code. If you are allowing more than one type of amino acid, simply list them with or without spaces or commas. Thus, SAV, S A V, or S,A,V will all specify serine, alanine, valine. The search is case insensitive.
- Use the Distance dropdown to select the distance (in Ångströms) to search within.
Add row for additional positions or click delete row to remove positions not required.
If the Chothia Position dropdown is not selected in a given row, no constraint is applied and the row is effectively removed from consideration. Similarly, if the at least one of option is selected in the Constraint dropdown, but nothing has been entered in the text box, no further constraint is applied.
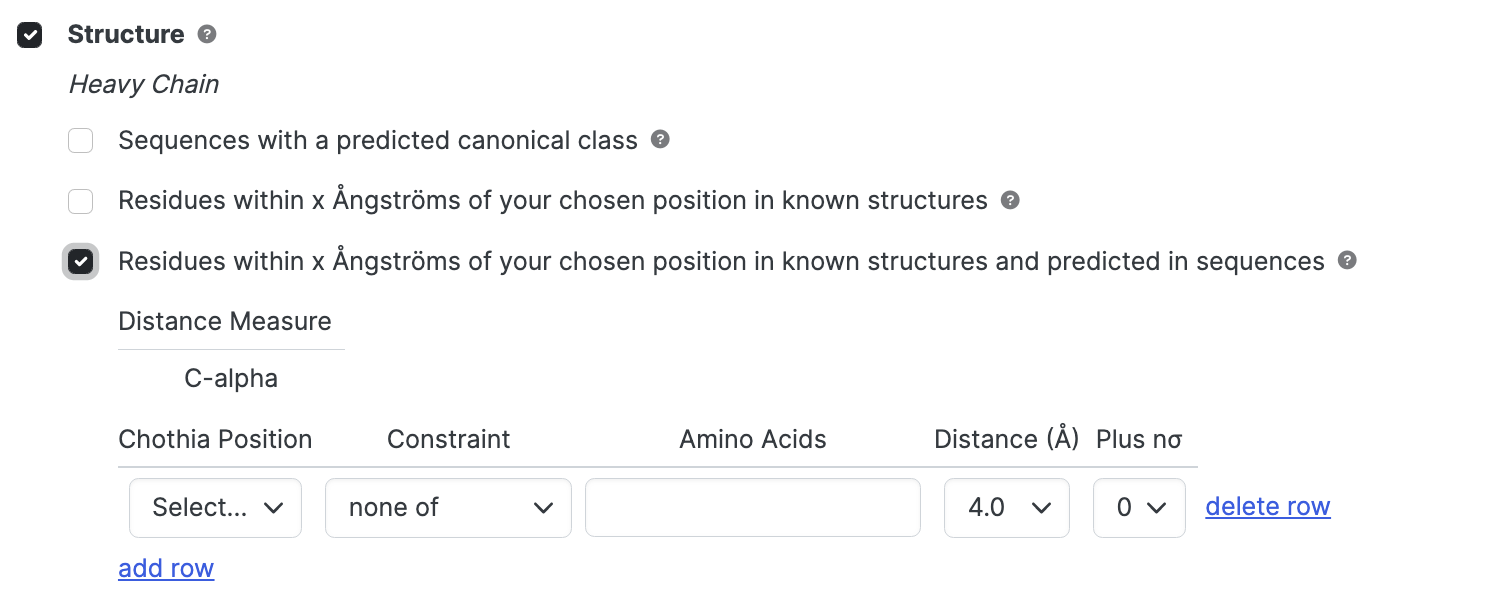
Residues predicted within X Ångströms; All sequences
 Note: This will search all entries using structural inference
Note: This will search all entries using structural inference
Restrict particular amino acid types within a particular distance of a specified residue including by inference sequences that are numbered but do not have structural data.
abYsis uses pre-calculated set of distance distributions calculated from known antibody structures to predict residues in the vicinity of a selected position based on correct numbering of the antibodies.
The original distributions were calculated using many hundreds of structures of numbered antibody chains so the prediction algorithm uses an average distance and a standard deviation.
- Distance Measure C-alpha to C-alpha distance between two residues
- Chothia Position selects required position.
- Constraint selects constraint to apply - either at least one of or none of. This controls whether the specified amino acids should be present or absent.
- Amino Acids Specify amino acids of interest using 1-letter code. To specifiy more than one type of amino acid, simply list them. You can choose with or without spaces or commas. SAV, S A V, or S,A,V will all work.
- Distance select mean distance (in Ångströms) between the two residues within which to search.
- Plus nσ select number of standard deviations to be added to the mean distance.
Results are brought back when m < d + nσ, where m the is mean C-alpha to C-alpha distance between the positions, d is the specified distance and σ is the standard deviation. Add row for additional positions or click delete row to remove positions not required.
If Chothia Position is not selected in a given row, no constraint is applied and the row is effectively removed from consideration.
If at least one of is selected in Constraint dropdown, but nothing has been entered in the text box, no constraint is applied.